Major milestone reached in effort to ID cancers’ genetic roots
Cancer Genome Atlas details genetic mutations driving cancer
 MATT MILLER
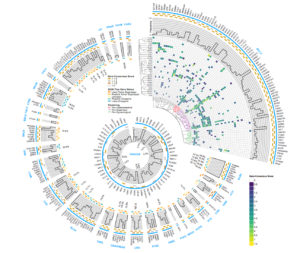
MATT MILLERLi Ding, PhD, led a team that analyzed DNA from more than 11,000 tumors across 33 cancer types. The project is part of The Cancer Genome Atlas, a national program aimed at understanding the genetic roots of cancer.
Researchers nationwide have reached a major milestone in describing the genetic landscape of cancer. Scientists at Washington University School of Medicine in St. Louis and other institutions have completed the genetic sequencing and analyses of more than 11,000 tumors from patients, spanning 33 types of cancer — all part of The Cancer Genome Atlas (TCGA) project, funded by the National Cancer Institute and National Human Genome Research Institute, both of the National Institutes of Health (NIH).
Altogether, the researchers identified about 300 genes that drive tumor growth. And, remarkably, just over half of all tumors analyzed carry genetic mutations that could be targeted by therapies already approved for use in patients.
TCGA was launched in 2005 to pursue the genetic basis of cancer. The results appear April 5 in a series of studies published in Cell Press journals.
“For the 10,000 tumors we analyzed, we now know — in detail — the inherited mutations driving cancer and the genetic errors that accumulate as people age, increasing the risk of cancer,” said leading TCGA scientist Li Ding, PhD, an associate professor of medicine and director of computational biology in the Division of Oncology at Washington University. “This is the first definitive summary of the genetics behind 33 major types of cancer.”
Ding, also an assistant director of The McDonnell Genome Institute at Washington University, is a major author of six papers published April 5 in the journals Cell, Cell Reports and Cell Systems, detailing the genetic mutations underlying cancer.
Genomic studies over the past decade have demonstrated that cancer is a disease of errors in genes rather than particular organs.
“This project is the culmination of more than a decade of groundbreaking work,” said NIH Director Francis S. Collins, MD, PhD. “This analysis provides cancer researchers with unprecedented understanding of how, where, and why tumors arise in humans, enabling better-informed clinical trials and future treatments.”
Ding said the new analyses have revealed that the genetic errors of cancer result in specific molecular signatures that could guide treatment.
“Rather than the organ of origin, we can now use molecular features to identify the cancer’s cell of origin,” Ding said. “We are looking at what genes are turned on in the tumor, and that brings us to a particular cell type. For example, squamous cell cancers can arise in the lung, bladder, cervix, and some tumors of the head and neck. We traditionally have treated cancers in these areas as completely different diseases. But studying their molecular features, we now know such cancers are closely related. Cancers originating in, for example, epithelial cells that line various organs are similarly closely related, regardless of their location.”
According to Ding, the research supports the idea that tumors of any type with high numbers of mutations — which often are resistant to chemotherapy — are susceptible to immunotherapy drugs called checkpoint inhibitors. Highly mutated tumors produce comparatively more misshapen proteins that can trigger an immune response. But as a safeguard against autoimmunity, the body often puts the breaks on such an immune response. Still, to treat aggressive tumors, checkpoint inhibitors can remove those breaks, letting the immune system fight the tumor more effectively.
The studies also provide further clarity on the significance of certain mutations in the BRCA1 gene that drive breast and ovarian cancer. Specific mutations in this gene are known to significantly increase the risk of certain types of cancer. But the consequences of many other mutations in this gene were unknown, making it difficult to predict cancer risk.
“We have known for a long time that BRCA1 is an important gene in cancer development,” said Ding, who has a research affiliation with Siteman Cancer Center at Barnes-Jewish Hospital and Washington University School of Medicine. “But it’s very hard to tease out which specific mutations in BRCA1 are actually driving the cancer and which mutations are harmless. Our paper on cancer-causing variants in inherited mutations provides new clarity on the BRCA1 mutations actually driving tumor growth. We found 21 disease-causing BRCA1 and BRCA2 variants in breast cancer, three in cervical cancer, one in colorectal cancer, one in glioblastoma and 38 in ovarian cancer.”
Ding said she is particularly excited about the prospect of using these analyses to re-examine data from past clinical trials. Many times, a small proportion of patients in a given trial did well on an experimental therapy, but many others did not respond to the treatment at all, and researchers didn’t understand why. Perhaps a drug was not approved for, say, lung cancer because of such results, but some patients with certain cancer mutations may benefit.
“Most earlier trials were not designed with genomics in mind,” Ding said. “We know how these patients responded. Now, we can sequence the tumor samples from patients enrolled in those trials with our latest software tools. We can look for correlations between the patients’ genomics and how they responded to the treatments. If we do this for many past trials, we will have tremendous statistical power to identify reasons why drugs work for some patients and not others. So even negative trials that might have been a disappointment at the time can become powerful tools to design better treatments in the future.”
In this way, a drug that might have failed as a treatment for lung cancer might be re-examined as a potential therapy for, say, squamous cell carcinoma, again, regardless of location.
“Even after genomic sequencing, sometimes we still can’t explain what is going on,” Ding said. “This is why we are planning to expand beyond studies of the tumor cells to include the entire tumor ecosystem — the immune cells that infiltrate the tumor and the supporting tissue that creates the tumor’s microenvironment.”
Added Eric Green, MD, PhD, director of the National Human Genome Research Institute, “These TCGA publications effectively showcase how genomics has brought transformative advances to the study of cancer. Central to these advances has been the development and use of new and increasingly more powerful technologies and approaches for genome sequencing.”
TCGA is a collaborative effort involving more than 20 institutions. The McDonnell Genome Institute and many researchers at Washington University have made major contributions to this national project. In the latest batch of seminal papers, Kuan-lin Huang, Matthew H. Bailey, Reyka G. Jayasinghe, Qingsong Gao, Song Cao, Wen-Wei Liang, Steven M. Foltz and others from Ding’s lab are primary authors. Within Washington University, Ding has collaborated with a number of investigators, including Feng Chen, PhD, an associate professor of medicine and a co-senior author of two of these papers.