
Kannampallil, Wiley named medical informatics fellows
Thomas Kannampallil, PhD, chief data scientist and assistant dean for data science at WashU Medicine, and Laura Wiley, PhD, an associate professor of neurology at WashU Medicine, were named 2025 American College of Medical Informatics (ACMI) fellows. They were among 24 professionals from the United States and abroad recognized by peers for their ongoing contributions to the field of data science.
Kannampallil, also an associate professor of anesthesiology, has extensive experience in the design, implementation and evaluation of novel AI-based technology designed to improve health care. He uses computer science and clinical informatics tools to implement applications at the point of care to help doctors make informed decisions. Kannampallil’s current research involves using electronic health record data to understand observable patient traits. In addition to his duties as the deputy editor for the Journal of Biomedical Informatics, Kannampallil serves on several national technical expert panels related to health information technology.
Wiley is an expert in computational phenotyping and has developed large electronic health record research repositories to study intracranial aneurysms and diabetes. She develops methods that use electronic health record data to generate clinical evidence and enhance precision medicine. Wiley previously served on the editorial board of the Journal of Biomedical Informatics and chaired the 2022 American Medical Informatics Association Informatics Summit. She has also led several courses on the use of secure electronic health record data for research.
ACMI is an honorary college of elected informatics fellows from the United States and abroad who have made significant and sustained contributions to the field of medical informatics and who have also met rigorous scholarly scrutiny. New fellows are elected annually by active members of ACMI. Their contributions to biomedicine and health care inform, educate and inspire the informatics community to advance human health.
Kannampallil and Wiley were honored and inducted as ACMI fellows during a ceremony at the 2025 American Medical Informatics Association annual symposium on Nov. 16 in Atlanta.
Nasal drops fight brain tumors noninvasively
Researchers at Washington University School of Medicine in St. Louis, along with collaborators at Northwestern University, have developed a noninvasive approach to treat one of the most aggressive and deadly brain cancers. Their technology uses precisely engineered structures assembled from nano-size materials to deliver potent tumor-fighting medicine to the brain through nasal drops. The novel delivery method is less invasive than similar treatments in development and was shown in mice to effectively treat glioblastoma by boosting the brain’s immune response.
The findings were published this month in PNAS.
Glioblastoma tumors form from brain cells called astrocytes and are the most common kind of brain cancer, affecting roughly three in 100,000 people in the U.S. Glioblastoma generally progresses very quickly and is almost always fatal. There are no curative treatments for the disease, in part because delivering medicines to the brain remains extremely challenging.
“We wanted to change this reality and develop a noninvasive treatment that activates the immune response to attack glioblastoma,” said Alexander H. Stegh, PhD, a professor and vice chair of research in the WashU Medicine Taylor Family Department of Neurosurgery and co-corresponding author of the study. Stegh also is research director of The Brain Tumor Center at Siteman Cancer Center, based at Barnes-Jewish Hospital and WashU Medicine. “With this research, we’ve shown that precisely engineered nanostructures, called spherical nucleic acids, can safely and effectively activate powerful immune pathways within the brain. This redefines how cancer immunotherapy can be achieved in otherwise difficult-to-access tumors.”
Cold tumors warmed with STING
Glioblastoma tumors are known as “cold tumors” because they do not induce the body’s natural immune response as do so-called “hot tumors” that are easier to treat with immunotherapies. Researchers have developed ways to spark an immune reaction against tumors by stimulating a pathway within cells called STING, which stands for stimulator of interferon genes. STING is triggered when a cell detects foreign DNA and activates the immune system to respond to the threat.
Past studies have shown that drugs activating STING in glioblastoma tumors can prime the body’s immune system to better fight the cancer. However, these agents break down quickly in the body and must be delivered directly into the tumor to work. Because repeated dosing is required for sustained benefit, relying on direct intratumoral administration requires highly invasive procedures.
“We really wanted to minimize patients having to go through that when they are already ill, and I thought that we could use the spherical nucleic acid platforms to deliver these drugs in a noninvasive way,” said Akanksha Mahajan, PhD, a postdoctoral research associate in Stegh’s lab and the first author on the study.
To overcome the problem, the Stegh team collaborated with co-corresponding author Chad A. Mirkin, PhD, director of the International Institute for Nanotechnology and the Rathmann Professor of Chemistry at Northwestern University, and his team. Mirkin invented spherical nucleic acids, a class of nanostructures that arrange DNA or RNA densely around a nanoparticle core, and he has shown that they have greater therapeutic potency compared to the standard delivery methods. The WashU Medicine and Northwestern researchers prepared a new class of spherical nucleic acids with gold cores studded with short snippets of DNA to trigger activation of the STING pathway in specific immune cells. To deliver these drugs to the brain, the team turned to the nose.
Intranasal therapy has been explored as a potential delivery method for medications targeting the brain, but no nanoscale therapies had yet been developed using this method to activate immune responses against brain cancers.
“This is the first time that it has been shown that we can increase immune cell activation in glioblastoma tumors when we deliver nanoscale therapeutics from the nose to the brain,” Mahajan said.
The team wanted to show that this approach could be used to deliver the medicine selectively to the brain, and that it would act on the appropriate cells once it got there. For the first objective, they used a molecular tag on the spherical nucleic acid that was visible under near-infrared light. They found that the nanomedicine, when delivered as droplets into the nasal passages of mice with glioblastoma, traveled along the path of the main nerve that connects facial muscles to the brain. The immune response evoked in the brain by the medicine was concentrated in the specific immune cells, especially those in the tumor itself, and triggered some helpful responses in the lymph nodes. The medicine did not spread to other parts of the body where it might cause unwanted side effects.
Examinations of immune cells in and near the tumor showed that the therapy successfully activated the STING pathway and armed the immune system to fight the tumor.
When applied in combination with drugs designed to help activate T lymphocytes, another type of immune cell, the new therapy eradicated the tumors with just one or two doses and induced long-term immunity against their recurrence. Taken together, the results were much better than those of current STING-activating immune therapies.
Stegh cautioned that firing up the STING pathway isn’t capable of curing glioblastomas without reinforcement from other therapeutic approaches. Turning on the STING pathway by itself isn’t enough to fight glioblastoma, because the tumor has many ways to block or shut down the immune response that STING is meant to activate. His team is looking to add capabilities to their nanostructure that activate other immune responses. This could allow physicians to double or triple the therapeutic targets all in a single therapy.
“This is an approach that offers hope for safer, more effective treatments for glioblastoma and potentially other immune treatment-resistant cancers, and it marks a critical step toward clinical application,” said Stegh.

From a Whisper to a Roar: Mellve’s Mission

Obituary: Brian Van Tine, professor of medicine, 53
Brian A. Van Tine, MD, PhD, a nationally recognized leader in sarcoma research at WashU Medicine, died Saturday, Nov. 8, 2025, at Barnes-Jewish Hospital following an acute illness. He was 53.
Van Tine was a professor of medicine and of pediatrics in the Department of Medicine’s Division of Oncology. He directed the Sarcoma Program at Siteman Cancer Center, based at Barnes-Jewish Hospital and WashU Medicine, where he built one of the country’s largest research and clinical programs in sarcoma, rare cancers of soft tissues that affect both adults and children. He treated adults and adolescents at Siteman and pediatric patients at Siteman Kids at St. Louis Children’s Hospital.
A highly accomplished and respected physician-scientist, Van Tine was a pioneer in research on tumor metabolism. He identified ways to exploit the unique metabolism of cancer cells to develop new therapies that kill sarcoma cells by cutting off their fuel supply while sparing healthy cells. He excelled at translating scientific discoveries made in his lab into clinical trials. He founded and chaired Siteman’s Sarcoma Tumor Board, a weekly meeting of cross-disciplinary experts who review individual patient cases and clinical trials.
In 2021, Van Tine was appointed director of developmental therapeutics for phase 1 clinical trials at Siteman. Under his leadership, the program expanded substantially and conducted many first-in-human clinical trials. He also served as the WashU Medicine principal investigator for more than 100 clinical trials, including multiple investigator-initiated clinical trials based on his lab’s research.
“Brian was not only a brilliant scientist but also a compassionate clinician and an extraordinary mentor,” said Victoria J. Fraser, MD, the Adolphus Busch Professor and head of the WashU Medicine Department of Medicine. “He dedicated his career to improving outcomes for patients with sarcoma. He shaped not only the field but also the lives of many patients and the careers of those fortunate to work alongside him. Patients and staff alike adored him for his warmth, candor and kindness.”
A native of Phoenix, Van Tine attended the University of Arizona, Tucson, where he earned dual bachelor’s degrees in chemistry and biochemistry in 1995. He continued his scientific training at the University of Alabama at Birmingham, where he earned a doctoral degree in pathology in 2003 and his medical degree in 2005. Later that year, he came to St. Louis and began specialized training in internal medicine and oncology, completing his internship, residency and fellowship — including serving as chief fellow in hematology and oncology — at WashU Medicine and Barnes-Jewish Hospital. In 2011, he joined the faculty of the Division of Oncology and was named director of the WashU Medicine Sarcoma Program.
“Brian’s work changed how we understand and treat sarcomas, and his influence will endure for decades to come,” said Daniel C. Link, MD, the Alan and Edith Wolff Endowed Professor, director of the Division of Oncology, and deputy director of Siteman. “He will be remembered for his boundless enthusiasm for scientific discovery, his rigorous intellect and his compassionate care of patients.”
Van Tine also was known as an inspirational mentor and educator. Many of the medical students, residents, fellows and junior faculty members he trained have gone on to successful careers in oncology and cancer research.
Van Tine was a central figure in national and international oncology organizations, where he served on advisory panels and committees for the National Cancer Institute of the National Institutes of Health (NIH), the American Association for Cancer Research, the American Society of Clinical Oncology and NRG Oncology. He helped organize many prestigious sarcoma conferences and was often an invited speaker at such events both in the U.S. and abroad. He also served on the editorial boards of influential cancer journals, including the Journal of Clinical Oncology, Clinical Cancer Research and the Annals of Oncology.
Van Tine is survived by his husband, Josh Hall; his mother, Carole Van Tine; his brother, Matthew Van Tine; and his mother-in-law, sisters-in-law, nieces and nephews.
Memorial contributions may be made to the Town and Country Symphony Orchestra.
Read more in the family obituary.
Why one man with a genetic predisposition for Alzheimer’s disease is defying the odds

Together, WashU Medicine MD, OT, PT students treat patients at Pro Bono Health Clinic
Makenna Dixon remembers all the appointments during her childhood.
Her mother suffered from autoimmune diseases that required countless trips to physical therapy. As her health conditions worsened, Dixon’s mom could no longer work a full-time job, and the lack of insurance through an employer made therapy one financial challenge among many.
For Dixon, now a third-year student in the WashU Medicine Program in Physical Therapy, watching her mother’s health decline fueled her fervor to advocate for people who don’t have health insurance. Those early experiences also influenced her decision to pursue a degree in physical therapy, and the Pro Bono Health Clinic drew her to study at WashU Medicine.
The WashU Medicine Pro Bono Health Clinic offers free occupational therapy (OT), physical therapy (PT) and medical intermediary services to uninsured community members in the St. Louis area. Medical intermediary care covers conditions such as diabetes, hypertension and other acute concerns for patients who are waiting to become established with a primary care provider.
The student-led effort, under direct clinical faculty supervision, provides informed, compassionate care to patients and trains learners through hands-on opportunities that introduce them to clinical and nonclinical factors that impact health.
Dixon said one day she hopes to manage a pro bono clinic of her own.
“Too often the community members we see struggle with basics like lack of food or transportation or a roof over their heads — things that can significantly impact their everyday lives and overall health,” Dixon said. “That’s why the Pro Bono Health Clinic prioritizes listening and addressing the whole patient.”
Patient-first, student-run
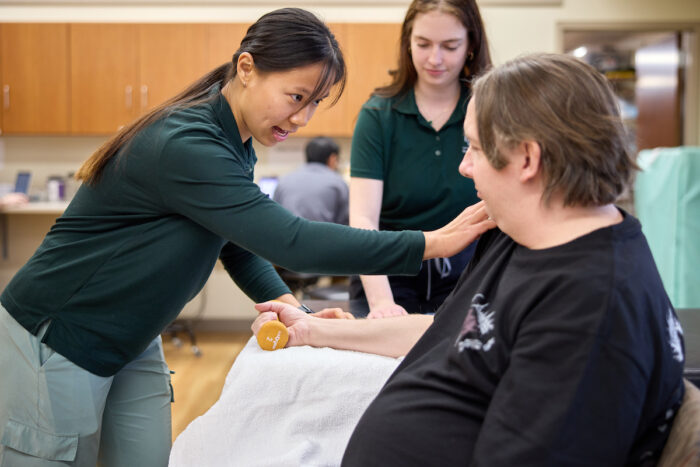
Each week, Dixon and a rotating team of other students see patients like Isheka Reynolds.
Reynolds, a St. Louis resident and mother of three, likens her ailments to a game of pingpong. The shots of pain start in her back, travel down her left hip and then send burning, tingling sensations to her knees and numbing toes.
“It’s like my body is trying to push and pull me all at the same time,” Reynolds said. Her herniated discs and neuropathy not only make it hard for Reynolds to get out of bed some days, but they also make it next to impossible to maintain a job.
The Pro Bono Health Clinic was designed with people like Reynolds in mind.
 Matt Miller / WashU Medicine
Matt Miller / WashU MedicineShe visits the clinic for physical therapy every other week, free of charge. By helping Reynolds with movement correction exercises and modifications to everyday positions and activities, students strive to alleviate her pain, bring a smile to her face and also check on any nonclinical needs she may have. Meanwhile, Stacy Tylka, physical therapy lead and a professor of physical therapy, oversees the treatment, willing to provide feedback as necessary.
 Matt Miller / WashU Medicine
Matt Miller / WashU MedicineTylka says the clinic was designed so that students serve as the primary caregivers while clinical professors take a more supervisory role. The environment offers learners the opportunity to refine their patient interviewing skills, practice building rapport, learn flexibility in real time and master the art of letting patients lead the discussion.
“Collectively, these experiences enable students to be adaptable and always put the patients’ needs first,” Tylka said.
Although Reynolds currently lives with her daughter, she hopes the pain will subside enough that she can work again and someday have a place of her own.
“I feel honored and thankful to be a patient at the Pro Bono Health Clinic,” she said. “I am very grateful to be in the care of such an amazing team.”
Collaboration across disciplines
Planning and design began long before the Pro Bono Health Clinic officially opened in February 2022. Tylka joined WashU Medicine colleagues Barbara Lutey, MD, the clinic’s medicine lead and an associate professor of medicine, and Jessica Dashner, occupational therapy lead and an associate professor of occupational therapy, to envision what this OT/PT/medical intermediary one-stop shop could become.
With no playbook to consult, the planning team worked together during the height of the COVID-19 pandemic to design a patient-centric model that was inclusive of all three disciplines, run by students and available to uninsured and underinsured St. Louisans each week.
“This type of endeavor would never happen without such a collaborative team,” Dashner said. “Each of us knew what our particular discipline could do to serve St. Louis community members, but it was truly transformative to merge these ideas into an end product that represented all of our perspectives and seamlessly addressed our patients’ and learners’ unique needs.”
The Pro Bono Health Clinic intentionally makes all three disciplines available from one centralized location. Sophia Dietz, a second-year medical student and student leader at the clinic, appreciates that individuals can have three caregivers sit and listen to their story all at the same time, so patients do not have to navigate multiple facilities and appointments.
“It’s been inspiring to be a part of a community of individuals who are all specialists in their area, who then work together with the patient to come up with the best solutions to improve their condition.”
 Matt Miller / WashU Medicine
Matt Miller / WashU MedicineBridging the gap
Studies show that up to 80% of a person’s health – including their physical environment, social and economic outlook, and health behaviors — may weigh more heavily on wellness than clinical care alone. That’s why the Pro Bono Health Clinic doesn’t stop at offering OT, PT and medical intermediary services. It also offers a bridge to other points of care, services and resources.
 Matt Miller / WashU Medicine
Matt Miller / WashU MedicineDepending on a patient’s needs, the medical intermediary team determines next steps for each individual. When needs surpass the scope of what the clinic can provide, it relies on a partnership with the St. Louis Integrated Health Network (IHN), which represents a majority of the region’s health care safety net. The IHN is a health care intermediary composed of four federally qualified health centers, two major hospital systems, two medical schools — including WashU Medicine — and two public health departments. Through its programs and initiatives, IHN and its partners collaborate to meet the health-care needs of the St. Louis region.
Cheryl Clerkly, an IHN community referral coordinator, works alongside the medical intermediary team most Fridays. Together they assess patients’ immediate needs and discuss relevant resources. Clerkly then assists patients with higher-level needs such as applying for public assistance, arranging reliable transportation for medical appointments or applying for housing.
Impacting lives and learners alike
A few months ago, Dixon had a memorable encounter at the Pro Bono Health Clinic, and she knew who she wanted to tell first.
“My patient had experienced a lot of mental health issues on top of mobility challenges,” Dixon said. “At the end of the session, she said it was one of the first times in her life that someone really listened to her and was willing to help.”
Dixon went straight home and called her mom.
“Even now it makes me want to cry tears of joy, thinking about how I not only helped the patient gain movement, but I was there for her in a way no one else was,” Dixon said. “Together we found resources that will change her life for the better.”
 Matt Miller / WashU Medicine
Matt Miller / WashU Medicine
Visit the Pro Bono Health Clinic website to learn more.

Inaugural WashU Medicine Dean’s Medals honor extraordinary contributions
Leaders across WashU Medicine foster a culture of ambition and achievement that improves the health of the region, the nation and the world. Some of these contributions leave such an indelible mark — advancing the institution’s mission in extraordinary ways — that they merit special recognition.
To honor such achievements — and the donors whose generosity makes them possible — David H. Perlmutter, MD, executive vice chancellor for medical affairs, the Spencer T. and Ann W. Olin Distinguished Professor and the George and Carol Bauer Dean of WashU Medicine, established a new award this year: the WashU Medicine Dean’s Medals.
“The Dean’s Medals were created to recognize extraordinary contributions that advance our mission — from those who treat patients and push the frontiers of science, to those who help lead the education and training of the next generation of physicians, to those whose advocacy and giving spirit shape the future of this institution,” Perlmutter said.
“Those receiving this honor are the ones who continually set that bar, who create and perpetuate and power our environment of never resting on laurels and always pushing forward.”
The following individuals received this prestigious award: Longtime WashU Medicine benefactors Debra and George Couch for leadership and service; Tasnim A. Najaf, MD, for clinical excellence; Eva Aagaard, MD, for education; Robert Schreiber, PhD, and Joan Luby, MD, for research; and Jeffrey Milbrandt, MD, PhD, and Aaron DiAntonio, MD, PhD, for innovation and commercialization.
Renée Shellhaas, MD, senior associate dean for faculty promotions and career development, and her team oversaw the awards process. “Congratulations to our outstanding recipients,” said Shellhaas, also the David T. Blasingame Professor of Neurology. “These medals celebrate remarkable people who lead with vision and courage — advancing discovery, elevating patient care, inspiring learners and driving innovation — all of which make WashU Medicine and our campus community truly exceptional.”
2025 WashU Medicine Dean’s Medal recipients

Debra and George Couch received the Dean’s Medal for Leadership and Service for their enduring financial support to Washington University and the research at WashU Medicine over the past four decades.
George Couch’s journey with WashU Medicine began in a deeply personal way. After his younger brother Gregory, who had dreamed of becoming a doctor, passed away unexpectedly in the 1980s, George and his family established the Gregory B. Couch Professorship in Psychiatry. That act of generosity set the stage for a lifetime of impact, touching countless patients and advancing WashU Medicine’s mission.
The Couches have continued to invest in the future of medicine locally, nationally and internationally. The Debra and George W. Couch III Biomedical Research Building — a hub for personalized medicine and discovery — stands as a testament to their ongoing commitment to giving, and they have been supporters of collaborative, interdisciplinary research within the building and across the university.

Tasnim Najaf, MD, a professor of pediatrics, received the Dean’s Medal for Clinical Excellence for developing three groundbreaking programs to improve care for young patients at St. Louis Children’s Hospital.
Najaf specializes in the care of critically ill newborns. As the medical director of the NICU extracorporeal membrane oxygenation (ECMO) program — which provides support to infants with critical heart and lung conditions — she leads a team of experts in ECMO, congenital diaphragmatic hernia and pulmonary hypertension. In addition, she has played an integral role in the Neonatal Renal Replacement Therapy Program, which provides lifesaving dialysis to infants. Najaf also helped implement the neonatal point of care ultrasound, which equips learners with bedside diagnostic and procedural ultrasound skills to diagnose various respiratory, cardiac and neurological conditions. These efforts have significantly improved outcomes for seriously ill babies.

Eva Aagaard, MD, the Carol B. and Jerome T. Loeb Professor of Medical Education, vice dean for education and vice chancellor for medical education, received the Dean’s Medal for Education for her visionary leadership in the development and implementation of the School of Medicine’s Gateway Curriculum.
Nationally known for her leadership in medical education, Aagaard re-envisioned the school’s MD educational program for the first time in more than two decades. Applying both personal and professional insights, Aagaard led the development of the Gateway Curriculum, which launched in 2020. The curriculum has increased learners’ awareness of health disparities, strengthened community engagement in the region and introduced more hands-on clinical experiences in the first two years of the MD program. She also oversees the school’s graduate medical education, allied health programs and PhD training programs. With Aagaard at the helm, WashU Medicine sets the standard as a world leader in medical education.

Robert Schreiber, PhD, the Andrew M. and Jane M. Bursky Distinguished Professor of Pathology & Immunology, received a Dean’s Medal for Research Excellence for his groundbreaking discoveries of the biological underpinnings of immune responses to cancer.
Cancer treatment once only consisted of surgery, radiation or chemotherapy. In recent years, more precise alternatives have emerged, thanks in large part to Schreiber’s internationally recognized pioneering research in cancer immunoediting. Schreiber serves as the founding director of the Bursky Center for Human Immunology & Immunotherapy, which translates human immunology research from the bench to the bedside. After helping unlock vital discoveries in how cancer evades the immune system, he went on to identify targets for immunotherapies. His ongoing work with cancer vaccines and immune-based treatments portends even greater impact for cancer patients in the future.

Joan Luby, MD, the Samuel and Mae S. Ludwig Chair in Psychiatry, received a Dean’s Medal for Research Excellence for her pioneering intervention to treat depression in early childhood as founder and director of the WashU Medicine Early Emotional Development Program.
Growing evidence underscores the mental health crisis taking place among children and adolescents across the country. Luby’s large-scale, internationally recognized studies have expanded the field’s understanding of the earliest manifestations of anxiety and depression and demonstrated that infants and children who struggle with emotions and behavior as early as age 3 are more vulnerable to depression and suicide later in life. In an effort to reverse this trend, her groundbreaking research continues to shed light on the importance of early identification and dyadic interventions that empower patients and their parents alike. Her novel early childhood parent-child psychotherapy approach has proven to yield dramatically improved outcomes for children that sustain into preadolescence.


Jeff Milbrandt, MD, PhD, the James S. McDonnell Professor of Genetics and executive director of the McDonnell Genome Institute, and Aaron DiAntonio, MD, PhD, the Alan A. and Edith L. Wolff Professor of Developmental Biology, received the Dean’s Medal for Innovation and Commercialization for their high-caliber scientific research and commercialization achievements.
Most neurodegenerative diseases such as ALS and peripheral neuropathy involve the early loss of axons — the fibers in the nervous system that are responsible for zapping electrical signals from one nerve cell to another. Blending Milbrandt’s research into the genomic and metabolic underpinnings of axon degeneration with DiAntonio’s research into the molecular mechanisms that control neural circuitry led to a surprising discovery: A molecule called SARM1 is responsible for the degeneration of axons. They showed this protein is an enzyme and that inhibiting this activity blocked axon loss and prevented disease. In short, their research uncovered the common molecular culprit found in most neurological disorders.
Working with WashU’s Office of Technology Management, the duo went on to found Disarm Therapeutics, where they developed therapies to inhibit the SARM1 pathway. In 2020, Eli Lilly and Company acquired Disarm Therapeutics for $135 million to speed development of new treatments for multiple neurodegenerative conditions. Milbrandt and DiAntonio have gone on to found several additional companies that focus on translating their research discoveries into therapeutic strategies to address unmet medical needs in neurodegenerative diseases. Their collaborative spirit exemplifies how WashU Medicine researchers can take a basic science discovery and transform it into a therapeutic advance that may one day help millions.
“Each of these honorees represents the very best of who we are and who we wish to be here at WashU Medicine,” Perlmutter said. “They embody the vision, the talent and the generosity of spirit that keep us at the forefront of medicine, science and education. Since this first Dean’s Medal ceremony is also my last as head of this exceptional institution, let me just say that I have considered it a great privilege to lead such an extraordinary group, and I look forward to watching from the sidelines as you soar to even greater heights.”

What Influencers and Critics Aren’t Telling You About Antidepressants
‘Light bulb moment’: New approach to treating Hep C postpartum a success in St. Louis

Promising clinical trials in Alzheimer’s prevention

Lang named Barbara J. Norton Professor of Physical Therapy
Catherine E. Lang, PhD, a leading researcher in the field of stroke recovery and rehabilitation in the WashU Medicine Program in Physical Therapy, has been installed as the inaugural Barbara J. Norton Professor of Physical Therapy. This is the program’s first named professorship, a milestone that signifies WashU Medicine’s commitment to groundbreaking research in the physical therapy field.
Lang’s research focuses on the development of effective, individualized rehabilitation for people recovering from stroke and other neurological injuries. She has established herself as a leader in the use of wearable movement sensors to quantitatively measure patients’ movement in daily life, helping to track their neurobehavioral recovery. Lang was installed by David H. Perlmutter, MD, executive vice chancellor for medical affairs, the Spencer T. and Ann W. Olin Distinguished Professor and the George and Carol Bauer Dean of WashU Medicine.
The professorship is named in honor of Barbara J. Norton, PhD, a WashU Medicine professor of physical therapy who has served on the faculty for more than 50 years — longer than any other faculty member in the program. Norton was instrumental in integrating research into the physical therapy curriculum.
“This professorship is a testament to Dr. Lang’s exceptional achievements in her research, teaching and practice,” said Chancellor Andrew D. Martin. “Establishing it in honor of Professor Barbara Norton, who has been instrumental in developing the Program in Physical Therapy into the teaching and research powerhouse it is today, serves as a fitting tribute to the program’s culture of excellence.”
Lang’s research seeks to clearly map the variables that are related to the speed and extent of neurobehavioral recovery after neurological injury. She has led numerous studies of interventions to increase the effectiveness of motor rehabilitation and to improve clinical outcomes, including first-in-human studies and early clinical trials. Lang’s research on wearable movement sensors has helped provide tools for practicing clinicians to streamline their assessments and develop informed prognoses.
“Dr. Lang has a tremendous track record of discoveries that have resulted in important advancements in patient care,” said Perlmutter. “Her work has improved the assessments that guide clinical decision-making and enriched the training and depth of knowledge available to students and practitioners in our physical and occupational therapy programs. Her commitment to research rigor, student training and improving patient outcomes embodies the best of what we do at WashU Medicine.”
Lang’s teaching and research have been recognized with several honors and awards, including the Academy of Neurologic Physical Therapy’s Excellence in Neurologic Physical Therapy Research Award and both the Marian Williams Award for sustained contributions to physical therapy research and the Helen Hislop Award for outstanding contributions to professional literature from the American Physical Therapy Association. She is a recipient of the NIH Merit Award from the National Center for Medical Rehabilitation Research and a fellow of both the American Society for Neurorehabilitation and the American Physical Therapy Association.
Her work is frequently cited in peer-reviewed research, and she has collaborated broadly both within WashU Medicine and with outside institutions. Her research partnerships span several disciplines, including biomedical engineering, neurosciences and social work, among others.
“I’ve watched the evolution of Dr. Lang’s work firsthand, and what has struck me is her ability to see the big picture and connect the dots in a way that nobody else can,” said Gammon Earhart, PhD, associate dean of physical therapy and director of the Program in Physical Therapy. “She is masterful at finding important questions to ask and exciting new directions to explore and then transforming them into clinically meaningful information that people can use in their practice.”
Lang earned her bachelor’s and master’s degrees from the University of Vermont and her PhD at WashU. She completed a postdoctoral followship at the University of Rochester in New York before returning to WashU Medicine in 2004 as a member of the faculty.
Norton’s time at WashU began when she was in high school and volunteered at the Irene Walter Johnson Institute of Rehabilitation, a medical rehabilitation facility that was merged into WashU Medicine in 1987. She earned her bachelor’s degree at WashU before joining the physical therapy program as an instructor. She then earned her master’s and doctoral degrees at WashU before becoming a professor. Norton was WashU Medicine’s first research physical therapist, conducting studies that sought to systematically measure and quantify spasticity.




